Shimamoto Group • Physics and Cell Biology Laboratory
Molecular and cellular biophysics dissecting cell division and development
Faculty
Research Summary
In most cells of our body, a variety of micron-sized structures, such as the nucleus and the mitotic spindle, assemble and function to control chromosome dynamics. Our laboratory uses advanced biophysical technologies, including intracellular micromanipulation, single-molecule imaging, and in vitro reconstitution, to visualize and manipulate such intracellular dynamics and unveil the intricated molecular mechanisms that ensure proper cell division and embryonic development.
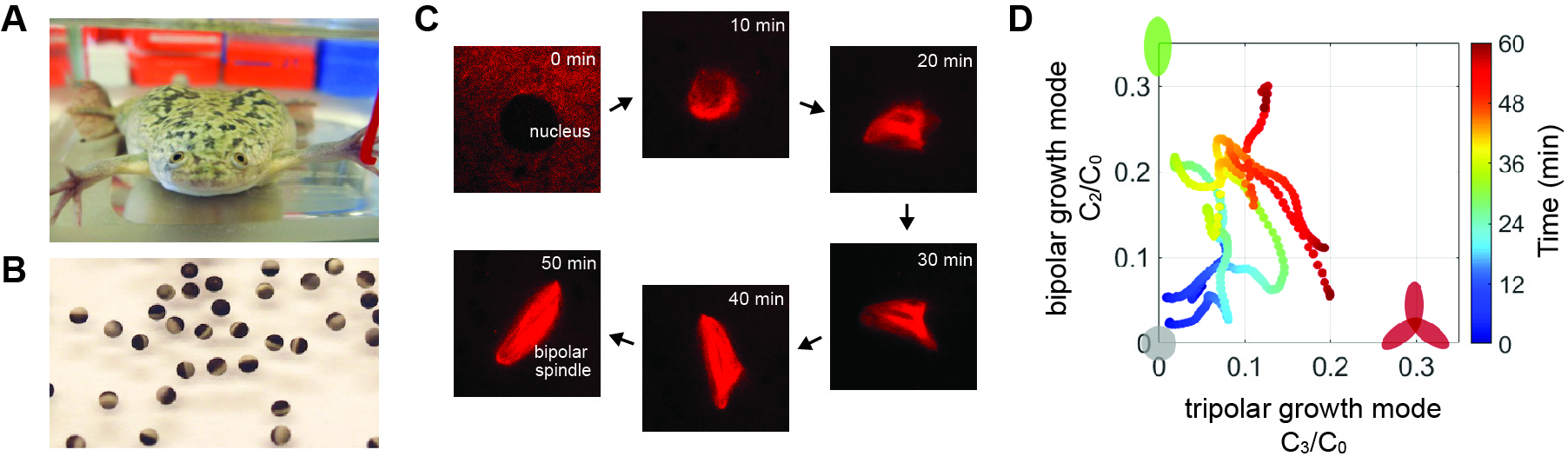
Selected Publications
Fukuyama T, Yan L, Tanaka M, Yamaoka M, Saito K, Ti SC, Liao CC, Hsia KC, Maeda YT, Shimamoto Y. Morphological growth dynamics, mechanical stability, and active microtubule mechanics underlying spindle self-organization. Proc Natl Acad Sci U S A. 2022 Nov;119(44):e2209053119.
Mori M, Yao T, Mishina T, Endoh H, Tanaka M, Yonezawa N, Shimamoto Y, Yonemura S, Yamagata K, Kitajima TS, Ikawa M. RanGTP and the actin cytoskeleton keep paternal and maternal chromosomes apart during fertilization. J Cell Biol. 2021 Oct 4;220(10):e202012001.
Takagi J, Sakamoto R, Shiratsuchi G, Maeda YT, Shimamoto Y. Mechanically Distinct Microtubule Arrays Determine the Length and Force Response of the Meiotic Spindle. Dev Cell. 2019 Apr 22;49(2):267-278.e5.

















